Abstract
-
Purpose
This study developed and validated a deep learning model for the automated early detection of androgenetic alopecia (AGA) using trichoscopic images, and evaluated the model’s diagnostic performance in a Korean clinical cohort.
-
Methods
We conducted a retrospective observational study using 318 trichoscopic scalp images labeled by board-certified dermatologists according to the Basic and Specific (BASP) system, collected at Ewha Womans University Medical Center between July 2018 and January 2024. The images were categorized as BASP 0 (no hair loss) or BASP 1–3 (early-stage hair loss). A ResNet-18 convolutional neural network, pretrained on ImageNet, was fine-tuned for binary classification. Internal validation was performed using stratified 5-fold cross-validation, and external validation was conducted through ensemble soft voting on a separate hold-out test set of 20 images. Model performance was measured by accuracy, precision, recall, F1-score, and area under the curve (AUC), with 95% confidence intervals (CIs) calculated for hold-out accuracy.
-
Results
Internal validation revealed robust model performance, with 4 out of 5 folds achieving an accuracy above 0.90 and an AUC above 0.93. In external validation on the hold-out test set, the ensemble model achieved an accuracy of 0.90 (95% CI, 0.77–1.03) and an AUC of 0.97, with perfect recall for early-stage hair loss. No missing data were present, and the model demonstrated stable convergence without requiring data augmentation.
-
Conclusion
This model demonstrated high accuracy and generalizability for detecting early-stage AGA from trichoscopic images, supporting its potential utility as a screening tool in clinical and teledermatology settings.
-
Keywords: Alopecia; Computer neural networks; Scalp; Deep learning; Dermatologists
Introduction
Background/rationale
Hair loss, especially androgenetic alopecia (AGA), is a common dermatological condition that has a considerable impact on patients’ quality of life. Early detection is essential, both for initiating timely treatment and for preventing further progression during the subtle and potentially reversible stages of the disease [
1]. In clinical settings, the Basic and Specific (BASP) classification system is widely used to assess the severity of hair loss, systematically categorizing frontal and vertex scalp patterns into structured scores [
2]. However, BASP scoring is based on manual visual assessment, which introduces subjectivity and variability between observers.
To overcome these limitations, deep learning–based approaches have increasingly been explored in dermatology, providing automated and objective tools for image-based diagnosis [
3]. Convolutional neural networks (CNNs), in particular, have shown strong performance in medical imaging tasks [
4], including trichoscopic image analysis [
5]. Building on this foundation, our study aimed to develop and validate a deep learning–based classification model capable of distinguishing BASP 0 (no hair loss) from BASP 1–3 (early-stage hair loss) directly from scalp images, with the goal of improving diagnostic reproducibility and standardization.
This study was specifically designed to address common challenges in medical image analysis, such as small dataset size and class imbalance. To evaluate model performance, we employed a 2-tier experimental strategy. First, we performed internal validation using stratified 5-fold cross-validation across the entire dataset to assess training stability and to identify optimal training configurations [
6]. Next, informed by these findings, we conducted external validation with a hold-out test set using ensemble voting, thereby simulating real-world application on previously unseen images [
7]. This sequential approach allowed us to evaluate both model training dynamics and real-world generalizability.
To support these experiments, we redefined BASP labels into binary categories (BASP 0 vs. BASP 1–3), implemented class-preserving validation through stratified sampling, and selected ResNet-18, a lightweight yet effective CNN architecture known for its balance of performance and computational efficiency in small- to medium-sized datasets.
Objectives
The aim of this study was to evaluate the model’s diagnostic accuracy, generalizability, and clinical utility using both internal cross-validation and external hold-out testing, providing evidence for its potential application in dermatological screening. Additionally, we sought to address class imbalance through stratified sampling, to assess the feasibility of binary BASP classification, and to demonstrate the use of a ResNet-18 CNN for the automated assessment of early-stage AGA.
Methods
Ethics statement
This study was approved by the Institutional Review Board (IRB) of Ewha Womans University Medical Center (IRB no., EUMC 2025-01-037). The requirement for informed consent was waived due to the retrospective nature of the study and the use of de-identified image data.
Study design
This was a retrospective observational study aimed at developing a deep learning model for classifying hair loss severity based on the BASP system.
Settings
A total of 318 trichoscopic images were collected from patients visiting the Department of Dermatology at Ewha Womans University Medical Center (Seoul, Republic of Korea) between July 7, 2018, and January 31, 2024. All images were acquired using the DermLite DL Cam Photo dermoscopy system (3Gen Inc.) and were captured from the frontal and vertex scalp regions during routine clinical assessments. Images of the occipital region, though used by dermatologists for clinical comparison during BASP scoring, were excluded from both model training and evaluation.
Participants
Eligible participants included patients aged 15 to 84 years who presented with concerns regarding hair loss. No additional inclusion or exclusion criteria were applied beyond clinical presentation, and all trichoscopic images with valid BASP annotations were included in the study. Labeling was performed by board-certified dermatologists using the BASP classification system, resulting in 151 images labeled as BASP 0 and 167 images labeled as BASP 1, 2, or 3 (
Table 1). There were no missing data in the final dataset used for model development and evaluation.
The primary outcome variable was binary classification of hair loss severity, defined as class 0 for BASP 0 and class 1 for BASP 1–3. This binary categorization was derived from the original 4-class BASP labels (BASP 0, 1, 2, and 3), which were assigned by dermatologists.
Image preprocessing and model configuration
Each trichoscopic image was paired with its corresponding BASP score using a consistent filename-label mapping system, enabling the model to learn the association between image features and hair loss severity. Images were resized to 224×224 pixels and normalized to a mean of 0.5 and standard deviation of 0.5 for each RGB channel.
A ResNet-18 CNN pretrained on ImageNet was used as the backbone. The final fully connected layer was replaced with a 2-unit output layer for binary classification. Model training was performed using the Adam optimizer (learning rate=0.001), a batch size of 128, and the cross-entropy loss function. No data augmentation or early stopping strategies were used. Each model was trained for a fixed 100 epochs.
Validation test
Internal validation: Stratified 5-fold cross-validation
The entire dataset (n=318) underwent stratified 5-fold cross-validation to ensure class balance within each fold. In every fold, 80% of the data were used for training and 20% for evaluation. The model with the highest validation accuracy over the 100 epochs was selected for reporting performance.
External validation: Hold-out ensemble testing
To evaluate generalizability, a hold-out test set of 20 images was created by randomly sampling 10 images from BASP 0 and 10 from BASP 1–3, ensuring class balance. The BASP 1–3 subset consisted of 4 BASP 1, 3 BASP 2, and 3 BASP 3 images, reflecting the distribution of hair loss stages (
Table 2). These 20 images were completely excluded from model training and validation processes.
The remaining 298 images were used to train 5 ResNet-18 models via stratified 5-fold cross-validation. Each model was then applied to the hold-out set. Final predictions were determined by ensemble soft voting, where the average class probabilities from the 5 models were combined to determine the predicted label. This ensemble approach was intended to simulate a real-world diagnostic scenario and enhance robustness in performance estimation.
Bias mitigation strategies
To address potential sources of bias in this small and imbalanced dataset, stratified k-fold cross-validation was used to ensure that all original samples were included in both training and validation while maintaining class distribution across folds. This approach mitigated selection bias and maximized data utility. Data augmentation was intentionally excluded to avoid introducing artificial variability. Additionally, ensemble prediction via soft voting across 5-fold-specific models was used in the hold-out test phase to reduce model variance and improve generalizability.
Study size
In total, 318 images were analyzed. No a priori sample size calculation was performed; instead, all eligible labeled images from the institutional database were used to reflect real-world clinical data availability. Post hoc 95% confidence intervals (CIs) were calculated for model accuracy in the hold-out set, based on the primary endpoint of binary classification performance.
Evaluation metrics
Performance on the hold-out set was assessed using ensemble accuracy, confusion matrix, receiver operating characteristic (ROC) curve analysis, area under the curve (AUC), and the 95% CI for accuracy. Classification metrics included accuracy, precision, recall, F1-score, and AUC. For the hold-out evaluation, a 95% CI for accuracy was computed using the Wald method.
Statistical methods
All statistical analyses and model training were performed using Python ver. 3.9 (
https://www.python.org/) and PyTorch ver. 1.12 (Meta) in the Google Colab environment. Image preprocessing, model definition, training, and evaluation were implemented using in-house PyTorch-based scripts. Visualization of results, including ROC curves and confusion matrices, was conducted using Matplotlib ver. 3.7 (Hunter). No statistical hypothesis testing (such as P-values) was conducted, as the focus was on classification performance and generalizability rather than group comparisons. Python code is available in
Supplement 1.
Results
Participants
A total of 318 trichoscopic scalp images were included for binary classification. Of these, 159 images were labeled as BASP 0 (no hair loss, class 0) and 159 images were labeled as BASP 1, 2, or 3 (early hair loss, class 1). No data were excluded, and all labeled images were used in both model training and evaluation.
Internal validation: stratified 5-fold cross-validation
The complete dataset (n=318) was used in stratified 5-fold cross-validation, ensuring equal class distribution within each fold. Each fold was trained for 100 epochs, with the model achieving the highest validation accuracy selected. The best epoch and corresponding performance metrics—accuracy, precision, recall, F1-score, and AUC—are summarized in
Table 3.
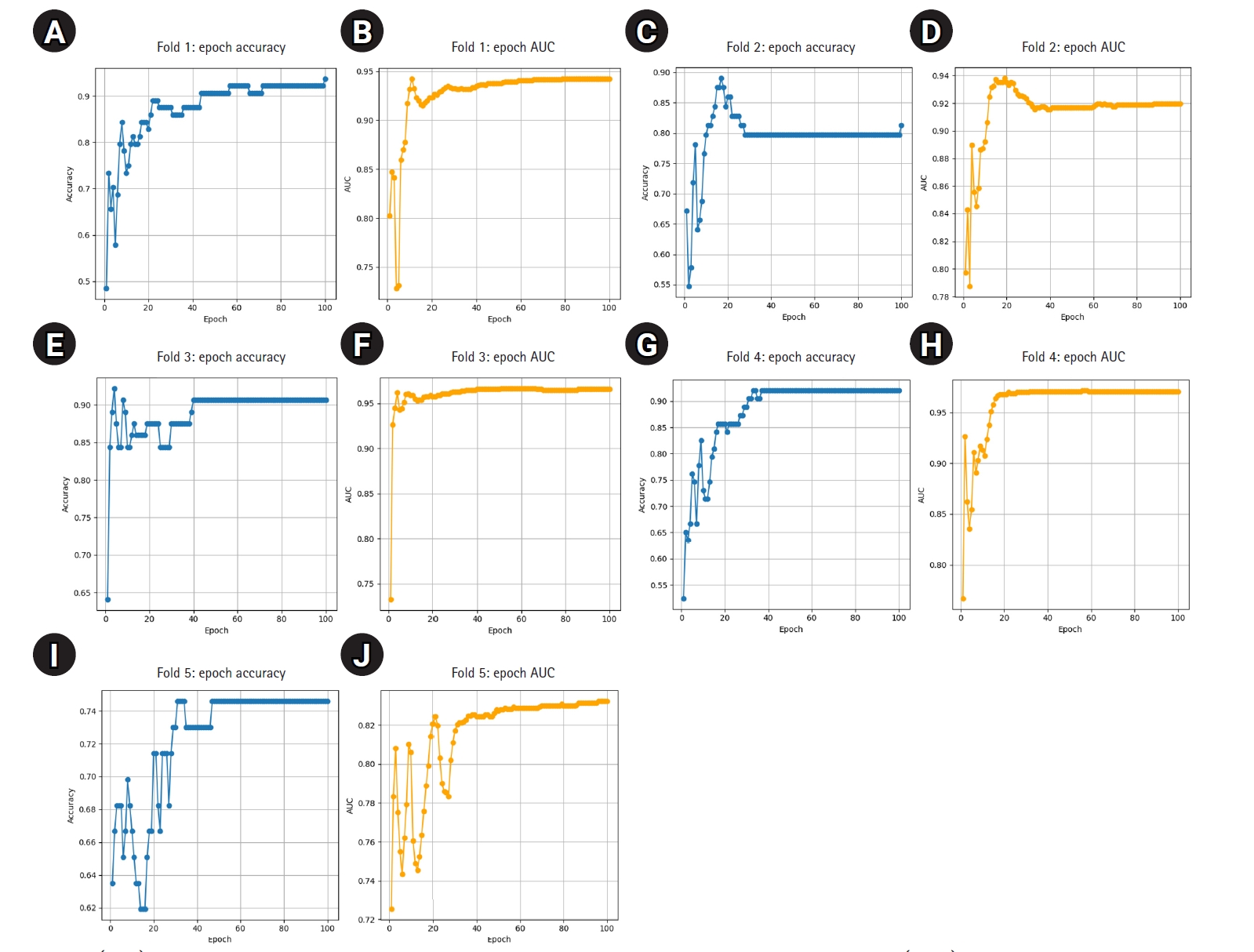
The model demonstrated stable performance across most folds, with 4 out of 5 achieving accuracy above 0.90 and AUC values above 0.93. One fold (Fold 5) showed relatively lower performance but still maintained an AUC of 0.8202. Detailed training curves for each fold are shown in
Fig. 1, illustrating how both accuracy and AUC progressed and converged over the 100 epochs. The average epoch time per fold ranged from 8.3 to 8.6 seconds, confirming the computational efficiency of ResNet-18 in this small- to medium-scale medical imaging task.
To assess generalizability, a separate hold-out test set of 20 images was created. The hold-out set included 10 class 0 and 10 class 1 images, selected via stratified sampling for balanced class representation. Detailed sample composition and individual model predictions are provided in
Supplement 2. These samples were excluded from training and reserved solely for external validation. The remaining 298 images were used to train 5 ResNet-18 models using stratified 5-fold cross-validation. The best-performing model for each fold was selected based on the highest classification accuracy on the respective test fold. These models, trained on the reduced dataset, demonstrated consistently high performance: all folds achieved accuracy above 0.80 and AUC above 0.86, with 1 fold reaching 0.95 in both metrics (
Table 4). These results indicate stable and effective training despite the reduced sample size. Detailed fold-wise metrics are presented below, and training curves are shown in
Fig. 2.
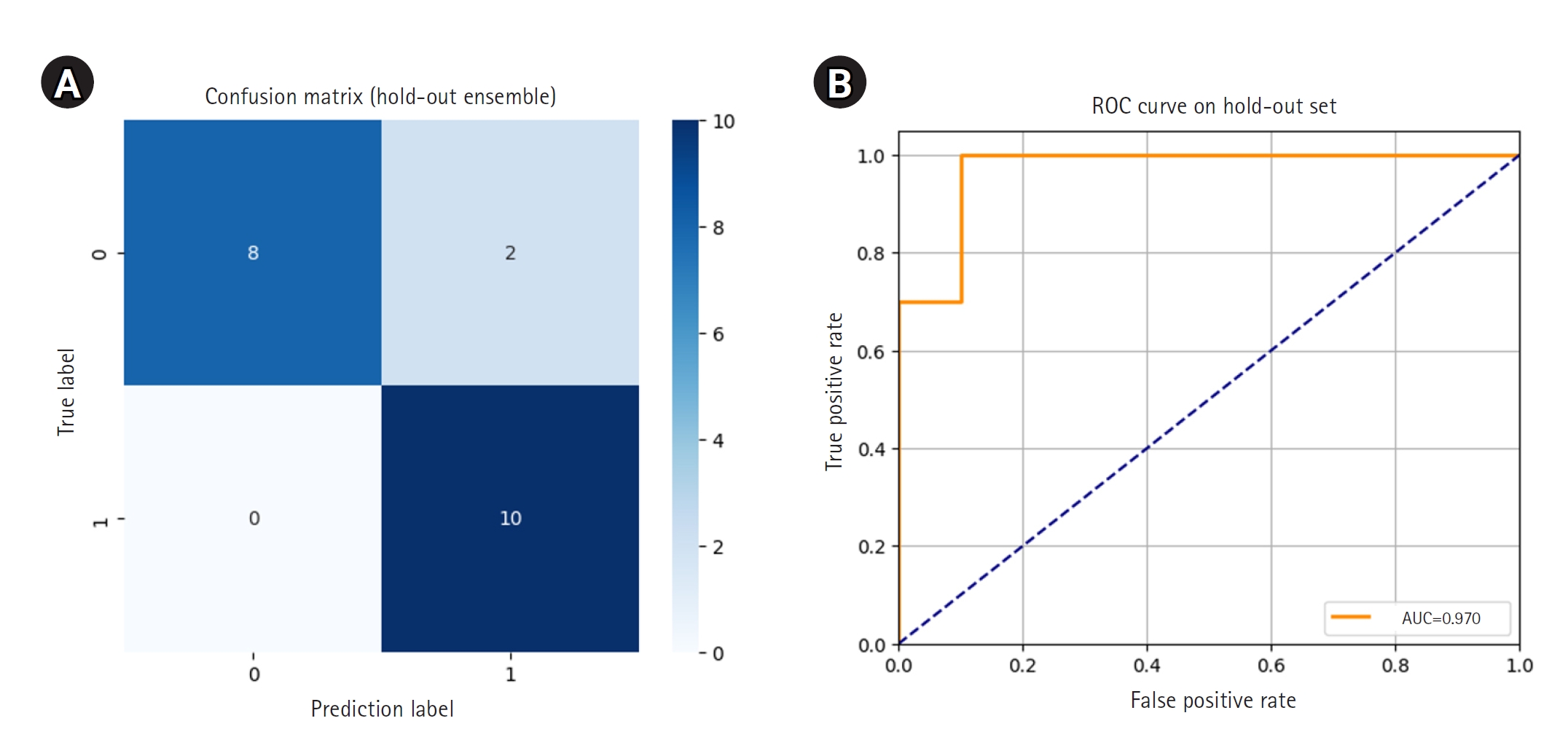
Each of the 5 models was then used to independently predict the 20-image hold-out test set. Final ensemble predictions were made using soft voting, averaging the predicted probabilities for each class across the 5 models. The ensemble model correctly classified 18 out of 20 images, achieving an accuracy of 0.9000. Notably, all 10 class 1 images were correctly identified, resulting in perfect recall for early hair loss detection. Two class 0 images were misclassified as class 1. The ROC curve showed an AUC of 0.970, and the 95% CI for accuracy, calculated using the Wald method, was 0.768 to 1.032.
The confusion matrix and ROC curve illustrating ensemble performance are presented in
Fig. 3. Detailed predictions for each of the 20 hold-out images, including model-specific votes and ensemble results, are provided in
Supplement 3.
Discussion
Key results
This study developed a ResNet-18-based deep learning model for classifying trichoscopic images by early-stage AGA severity using the BASP system. The model consistently demonstrated high performance in internal validation via stratified 5-fold cross-validation and maintained robust generalizability in external ensemble testing. High accuracy and AUC values in both validation settings confirmed the model’s reliable discrimination between BASP 0 and BASP 1–3, even on previously unseen data.
Interpretation
The goal of this study was to assess the feasibility of deep learning for early-stage hair loss screening based on the BASP classification. The model showed consistently high performance across stratified internal folds, reflecting its ability to learn relevant patterns from trichoscopic images despite the limited data. Notably, 4 out of 5 internal folds achieved strong accuracy and AUC scores, with only 1 fold showing relatively reduced performance, likely due to incidental variation in class composition within that particular split. Nevertheless, this fold still maintained a respectable AUC of 0.8202, suggesting overall robustness across validation subsets.
This stability may be attributed to the binary simplification of BASP labels (BASP 0 vs. BASP 1–3), which reduced class fragmentation and enhanced the signal-to-noise ratio during training. Stratified 5-fold cross-validation further mitigated bias from class imbalance and ensured that every image contributed to both training and validation—a critical design choice for small datasets. Importantly, data augmentation was deliberately excluded, yet the model still exhibited stable convergence across folds (
Figs. 1,
2), indicating that core patterns were sufficiently learnable from raw image features alone.
Performance on the external hold-out test set further validated the model’s generalizability. Ensemble soft voting, based on 5 independently trained models, successfully classified 18 out of 20 images, achieving 90% accuracy and an AUC of 0.970. All early hair loss cases (class 1) were correctly identified, resulting in perfect recall. In clinical screening, such a low false-negative rate is crucial for timely intervention and minimizing missed diagnoses.
These findings collectively suggest that, with careful design, such as label restructuring, stratified sampling, and ensemble evaluation, even small, real-world clinical datasets can support the development of reliable deep learning models for early-stage disease detection. While the external test set was limited to 20 images, the ensemble approach helped compensate for this limitation by reducing model variance and strengthening prediction confidence.
Comparison with previous studies
While previous research has applied CNNs to dermatologic imaging, few studies have specifically targeted early-stage AGA or integrated BASP classification into model development. Most existing approaches rely on multiclass classification or segmentation tasks, which typically require larger datasets and extensive manual labeling.
In contrast, this study demonstrates that clinically meaningful classification can be achieved through strategic label simplification and robust validation strategies, even with limited data. By employing stratified cross-validation and ensemble-based external testing, this work addresses a common gap in medical artificial intelligence (AI) research: the reliance on internal metrics without assessment of generalizability to unseen data.
Limitations
The main limitation of this study is its small sample size (n=318), which may restrict generalizability. In particular, images labeled as BASP 2 and 3 were underrepresented, potentially limiting the model’s ability to learn nuanced patterns across progressive hair loss stages. A larger, more diverse dataset would support more robust training and enable a more comprehensive evaluation, including an expanded hold-out test set. All images were obtained from a single institution under specific imaging conditions, which may not capture the full variability seen in wider clinical settings, such as differences in scalp types, lighting, or trichoscopic equipment. Additionally, only one CNN architecture (ResNet-18) was evaluated. Comparative assessments across multiple architectures or training configurations could provide greater insight into model optimization.
Training hyperparameters, such as batch size and learning rate, were not systematically optimized. Although a batch size of 8 was initially tested, training did not converge effectively under that condition. A batch size of 128 was subsequently adopted based on successful convergence and was maintained throughout the study. Beyond this adjustment, no systematic exploration or substudy was conducted to identify optimal configurations for further improving model performance in this dataset.
Finally, the performance variability observed across cross-validation folds—most notably in Fold 5—highlights the model’s sensitivity to class composition and sampling variation, underscoring the challenges of training on limited, imbalanced medical datasets.
Generalizability
Despite the limited dataset, ensemble-based hold-out testing demonstrated strong generalizability to unseen images. The model’s high recall for early-stage AGA suggests potential clinical value as a screening tool, especially in resource-limited settings such as primary care or teledermatology. However, because all data were sourced from a single institution using one specific dermoscopy device, and all images were from Korean patients, broader generalization across different populations (varying in age, sex, ethnicity, and imaging equipment) will require future validation using multicenter, multi-ethnic, and multi-device datasets.
Suggestions for further studies
To build upon these findings and develop a practical diagnostic framework for early-stage AGA, several future directions are suggested.
First, multicenter data collection encompassing diverse populations, imaging devices, and clinical environments is essential to enhance generalizability and reduce bias related to demographics or equipment. Larger and more balanced datasets would also enable finer label granularity—for example, distinguishing BASP 1 (early) from BASP 2–3 (progressive)—to better reflect the clinical spectrum of hair loss.
Second, comparative evaluation of alternative neural network architectures, such as EfficientNet, DenseNet, or vision transformers, should be performed to identify optimal trade-offs among diagnostic accuracy, computational efficiency, and deployment feasibility.
Third, integrating explainability techniques (such as Grad-CAM) and uncertainty quantification methods (like CIs or Monte Carlo dropout) may improve clinical trust and facilitate human–AI collaboration. Fairness metrics should also be monitored to assess potential bias across age, sex, or ethnicity subgroups.
Finally, real-world implementation studies in primary care or teledermatology—including workflow simulations and user feedback—will be vital for validating the model’s practical utility and educational value in early diagnosis scenarios.
Conclusion
This study demonstrated the feasibility of a deep learning–based approach for early detection of AGA by leveraging BASP score classification. By simplifying the task to a binary distinction between non-hair loss (BASP 0) and early hair loss (BASP 1–3), the model achieved strong performance in both internal validation and ensemble-based external testing, without requiring data augmentation or extensive hyperparameter tuning. The use of stratified cross-validation and ensemble soft voting enabled robust learning even with a limited dataset, suggesting practical applicability in clinical screening scenarios. In particular, the high recall for early hair loss cases indicates strong potential for timely intervention. These results support the integration of automated BASP-based assessment tools into dermatological workflows, promoting more standardized and objective evaluation of hair loss in clinical and teledermatology practice.
Furthermore, such deep learning–based systems may reduce the burden of manual labeling, minimize subjectivity in early hair loss diagnosis, and offer consistent alerts for possible AGA, thereby improving the accessibility, accuracy, and standardization of dermatologic care.
-
Authors’ contribution
Conceptualization: MJS, SHA, JYB. Data curation: MJS, JYB. Formal analysis: MJS. Funding acquisition: SHA, JYB. Methodology: MJS. Project administration: SHA. Visualization: MJS. Investigation: MJS. Resources: JYB, SHA. Software: MJS. Supervision: SHA, JYB. Writing–original draft: MJS. Writing–review & editing: MJS, SHA, JYB.
-
Conflict of interest
So Hyun Ahn has been an assistant editor of the journal since 2024, and Ji Yeon Byun has been an assistant editor of the journal since 2016. However, they were not involved in the editorial or peer-review process of this manuscript. Otherwise, no potential conflict of interest relevant to this article was reported.
-
Funding
None.
-
Data availability
Due to privacy concerns, the raw trichoscopic image data cannot be publicly shared (see Fig. 4 for a representative example).
-
Acknowledgments
None.
Supplementary materials
Fig. 1.(A–J) Epoch-wise accuracy and area under the receiver operating characteristic curve (AUC) for each fold.

Fig. 2.(A–J) Epoch-wise accuracy and area under the receiver operating characteristic curve (AUC) for each fold trained on 298 images.

Fig. 3.Confusion matrix (A) and receiver operating characteristic (ROC) curve (B) for ensemble predictions on the hold-out set. AUC, area under the ROC curve.

Fig. 4.(A, B) Example of scalp image and Basic and Specific (BASP) classification data.

Table 1.Distribution of original data
|
BASP label |
0 |
1 |
2 |
3 |
Total |
|
No. of images |
151 |
109 |
47 |
11 |
318 |
Table 2.Dataset configuration for hold-out ensemble voting test
|
Class 0 |
Class 1 |
|
BASP label |
0 |
1 |
2 |
3 |
|
Hold-out test set |
10 |
4 |
3 |
3 |
Table 3.Fold-wise metrics for stratified 5-fold cross-validation
|
Fold |
Best epoch |
Accuracy |
Precision |
Recall |
F1-score |
AUC |
|
1 |
100 |
0.9375 |
0.9677 |
0.9091 |
0.9375 |
0.9423 |
|
2 |
17 |
0.8906 |
0.8462 |
0.9706 |
0.9041 |
0.9353 |
|
3 |
4 |
0.9219 |
0.9143 |
0.9412 |
0.9275 |
0.9618 |
|
4 |
33 |
0.9206 |
0.9375 |
0.9091 |
0.9231 |
0.9707 |
|
5 |
31 |
0.7460 |
0.7429 |
0.7879 |
0.7647 |
0.8202 |
Table 4.Fold-wise metrics from training on the 298-image dataset
|
Fold |
Best epoch |
Accuracy |
Precision |
Recall |
F1-score |
AUC |
|
1 |
11 |
0.8333 |
0.8621 |
0.8065 |
0.8333 |
0.8788 |
|
2 |
14 |
0.8000 |
0.7632 |
0.9063 |
0.8286 |
0.8650 |
|
3 |
23 |
0.9500 |
0.9143 |
1.0000 |
0.9552 |
0.9542 |
|
4 |
22 |
0.8475 |
0.8929 |
0.8065 |
0.8475 |
0.9182 |
|
5 |
23 |
0.8644 |
0.8966 |
0.8387 |
0.8667 |
0.9389 |
References
- 1. Starace M, Orlando G, Alessandrini A, Piraccini BM. Female androgenetic alopecia: an update on diagnosis and management. Am J Clin Dermatol 2020;21:69-84. https://doi.org/10.1007/s40257-019-00479-x
- 2. Lee JY, Kim CH, Lee WS. Relationship between illness behavior and hair loss pattern according to the basic and specific (BASP) classification. Ann Dermatol 2023;35:318-320. https://doi.org/10.5021/ad.21.085
- 3. Lee S, Lee JW, Choe SJ, Yang S, Koh SB, Ahn YS, Lee WS. Clinically applicable deep learning framework for measurement of the extent of hair loss in patients with alopecia areata. JAMA Dermatol 2020;156:1018-1020. https://doi.org/10.1001/jamadermatol.2020.2188
- 4. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-118. https://doi.org/10.1038/nature21056
- 5. Kuhn M, Johnson K. Applied predictive modeling. Springer; 2013. https://doi.org/10.1007/978-1-4614-6849-3
- 6. Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
- 7. Ismail Fawaz H, Forestier G, Weber J, Idoumghar L, Muller PA. Deep learning for time series classification: a review. Data Min Knowl Discov 2019;33:917-963. https://doi.org/10.1007/s10618-019-00619-1